Introduction to Theory, Practice of Radioligand Binding
Radioactivity, Radioligands and Binding Assays
As we discussed, a radioligand is a radioactively labeled drug that
can associate with a receptor, transporter, enzyme, or any protein of interest. Measuring the rate and extent of binding provides
information on the number of binding sites, and their affinity and pharmacological (biological) characteristics.
There are three commonly used experimental protocols:
- Saturation binding experiments. These are the experiments
whose theoretical basis we just derived. In these experiments the extent of binding is measured in the presence of different
concentrations of the radioligand. From an analysis of the relationship between binding and ligand concentration, we can determine
the number of binding sites, Bmax, and ligand affinity, KD.
- Kinetic experiments. Saturation and competition experiments
are allowed to incubate until binding has reached equilibrium. Kinetic experiments measure the time course of binding and
dissociation to determine the rate constants for radioligand binding and dissociation. Together, these values also permit
a calculation of the KD.
- Competitive binding experiments. Measure the binding
of a single concentration of radioligand at various concentrations of an unlabeled competitor. Analyze these data to learn
the affinity of the receptor for the competitor.
How to Separate Bound from Free
For any of these approaches to work, one must be able to determine
how much radioligand is associated with the receptor, and how much is unbound. It is fortunate that the huge majority of target
macromolecules are either insoluble (e.g., membranous) or can be made insoluble with simple biochemical tricks (e.g., using
polyethylene glycol). There are three general approaches of which you should be aware.
Equilibrium dialysis
Technical problems prevent its use except in rare circumstances.
Some issues include: degradation or sticking of receptor or ligand; cumbersome nature of assays when large numbers of samples
are needed; time to obtain equilibrium; etc. Moreover, this technique cannot be used for kinetic analysis.
Centrifugation:
|
The [R] changes during pelleting and trapped radioactivity increases
non-specific binding (NSB; see later sections). One can calculate time allowable for separation based on KD and
the derived rate constants; the relationship will be logarithmic. The table to the left makes the assumption that separation
must be complete in ca. 0.15 t0.5 if one is to avoid losing more than 10% of DR complex, thus introducing unacceptable
error. As one can see, assays with low affinity ligands introduce very specific experimental problems.
|
KD (nM)
1000
100
10
1
0.1 |
0.15(t0.5) (sec)
0.1
1.
10.
100.
1000. |
Filtration:
With sufficiently high KD, several washes allow very
low NSB. Cold wash buffer (like a cold centrifuge above) will increase further the separation time [see values for centrifugation
(above)].
Total vs. free concentrations of ligand. Ligand depletion.
The equations that describe the law of mass action include the variable
F ([Ligand]), the free radioligand. In many experimental situations, you can assume that only a very small fraction
of the ligand ever binds to receptors. In these situations, you can assume that the free concentration of ligand is approximately
equal to the concentration you added. This assumption vastly simplifies the analysis of binding experiments, and the standard
analysis methods depend on this assumption. In other situations, a large fraction of the ligand binds to the receptors. This
means that the concentration of ligand free in solution does not equal the concentration you added, and the discrepancy is
not the same in all tubes or at all times. The free ligand concentration is depleted by binding. Many investigators
use this rule of thumb. If less than 10% of the ligand binds to receptors, don't worry about ligand depletion. If more than
10% of the ligand binds, you have three choices:
- Change the experimental conditions. The simplest approach is to
decrease the amount of receptor in the incubation by using less tissue or fewer cells. The problem is that this will decrease
the number of radioactive counts. The way around this is to increase the reaction volume without changing the amount of tissue.
The problem with this approach is that it requires more radioligand, which is usually very expensive.
- Measure the free concentration of ligand in every tube. This is
fairly straightforward if you use centrifugation or equilibrium dialysis, but is difficult if you use vacuum filtration. One
can also estimate the free concentration by subtracting the total bound from the total added.
- Use analysis techniques that adjust for the difference between
the concentration of added ligand and the concentration of free ligand.
Radioactivity
Specific Radioactivity
When you buy radioligands, the packaging usually states the specific
radioactivity as Curies per millimole (Ci/mmol). Since you measure counts per minute (cpm) , the specific radioactivity is
more useful when stated in terms of cpm. Often the specific radioactivity is expressed as cpm/fmol (1 fmol = 10-15
mole). To convert from Ci/mol to cpm/fmol, you need to know the efficiency of your counter. Efficiency is the fraction of
the radioactive disintegration that are detected by the counter.
Radionuclides that decay with high energy can be counted more efficiently
than those with low energies modes. For example, 125I can be counted at very high efficiencies, usually 70-90+%
depending on the geometry of the gamma counter (e.g., if the detector doesn’t entirely surround the tube, some gamma
rays miss the detector).
With 3H, the efficiency of counting is much lower (maximally
ca. 60%). The low efficiency is mostly a consequence of the physics of decay, and can not be improved by better instrumentation.
When a tritium atom decays, a neutron converts to a proton and the reaction shoots off an electron and neutrino. The energy
released is always the same, but it is randomly partitioned between the neutrino (not detected) and an electron (that we try
to detect). When the electron has sufficient energy, it will travel far enough to encounter a fluor molecule in the scintillation
fluid. This fluid amplifies the signal and gives off a flash of light detected by the scintillation counter. The intensity
of the flash (number of photons) is proportional to the energy of the electron. If the electron has insufficient energy, it
is not captured by the fluor and is not detected. If it has low energy, it is captured but the light flash has few photons
and is not detected by the instrument. Since the decay of many tritium atoms does not lead to a detectable number of photons,
the efficiency of counting is less than 100%.
To convert from Ci/mmol to cpm/fmol, you need to know that 1 Ci
equals 2.22 x 1012 dpm (disintegrations per minute). Another unit of radioactivity that is becoming more common
is the Becquerel (Bq). 1 mCi = 27.0270 MBq (that’s mega, not milli). The same exercise applies if you receive your radioactive
sample in units of Bq.
Use this equation to convert Z Ci/mmol to Y cpm/fmol
when the counter has an efficiency (expressed as a fraction) equal to E.
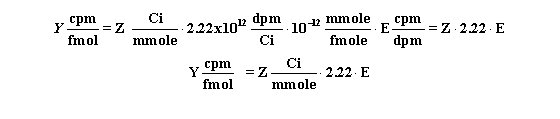
Calculating the concentration of the radioligand
Rather than trust your dilutions, you can accurately calculate the
concentration of radioligand in a stock solution. Measure the number of counts per minute in a small volume of solution and
use this equation. C is cpm counted, V is volume of the solution you counted in ml, and Y is the specific activity of the
radioligand in cpm/fmol (calculated in the previous section).
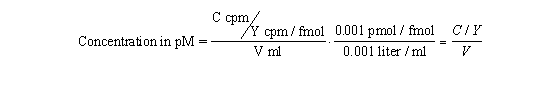
Radioactive decay
Radioactive decay is entirely random. A particular atom has no idea
how old it is, and can decay at any time. The probability of decay at any particular interval is the same as the probability
of decay at any other interval. If you start with N0 radioactive molecules, the number remaining at time t is:
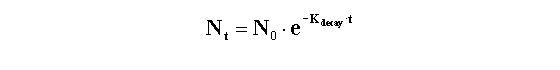
KDecay is the rate constant of decay expressed in units
of inverse time. Each radioactive isotope has a different value of KDecay. The half-life (t½) is the
time it takes for half the isotope to decay. Half-life and decay rate constant are related by this equation:

It is this relationship that allowed us to formulate the equation
presented earlier (and shown below) that uses the t0.5 rather than the less commonly seen Kdecay.
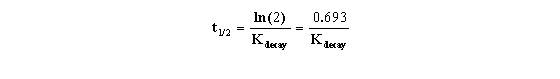
Selecting the radioligand
What chemical structure: agonist vs. antagonist
In general antagonists are used much more widely, in large measure
because they often have much higher affinity than available agonists. (Discussion: why is this?) Resulting technical problems
(degradation or sticking of receptor or ligand; cumbersome nature of assays when large numbers of samples are need; time to
obtain equilibrium; etc.) have limited use of agonists with most receptors.
Choice of isotope
One can predict what type of radionuclide can be used successfully
based on the density of receptors in the preparation being studied. This is either known from direct experimental evidence,
or can be estimated by analogy to well characterized systems. Usually, one finds densities of from 10-500 fmol receptor/mg
protein in "normal neural tissue", and from 200-3,000 fmol/mg in transfected cells, depending on the promoter used and other
factors.
One can decide what radionuclides are suitable based on the
estimated density of receptors and on elementary principles of detection of radioactivity. These calculations needed
to determine feasibility for use of a radioligand in radioreceptor assays are identical to those for the use of radioisotopes
in any biology problem. If you are not familiar with such calculations, please ask in class or see me before class.
The factors to consider include:
- environmental and experimental background
- efficiency of counting
- amount of radiolabel incorporation into radioligand
- specific activity of radionuclide
The specific activity of the radionuclide is based solely on
its half-life, and is independent on the mode or energy of decay. The following table shows the half-lives for commonly used
radioisotopes. The table also shows the specific activity assuming that each molecule is labeled with one atom of an isotope
(as is often the case with 125I and 32P). Tritiated molecules often incorporate several tritium atoms,
resulting in increased the specific radioactivity of the molecule.
| Radionuclide |
Half life |
Specific Activity (Ci/mmol) |
Decays to: |
b Energy (keV) |
| 3H |
12.43 y |
28.8 |
3He |
18 |
| 125I |
59.6 d |
2176 |
125Te |
- |
| 32P |
14.3 d |
9131 |
32S |
1710 |
| 35S |
87.4 d |
1494 |
35Cl |
167 |
| 14C |
5730 y |
0.062 |
14N |
156 |
You can calculate radioactive decay from a date where you knew the
concentration and specific radioactivity using this equation (see later sections for more detail).

It turns out that the decay of most isotopes of biological interest
result in either destruction of the molecule in which the atom is contained or in significant chemical change (see table;
for example 32P decays to 32S). Thus, rather than changing the specific radioactivity of the ligand
(as is commonly - and mistakenly - done), the concentration of radioligand is reduced.
The Poisson distribution
The decay of a population radioactive atoms is random, and therefore
subject to a sampling error. (This sampling error has nothing to do with other experimental factors, such as the differences
in efficiency of counting between samples.) For example, the radioactive atoms in a tube containing 1000 cpm of radioactivity
won’t give off exactly 1000 counts in every minute. There will be more counts in some minutes and fewer in others, with
the distribution of counts following a Poisson distribution.
After counting a certain number of counts in your tube, you want
to know what the "real" number of counts is. Obviously, there is no way to know that. But you can calculate a range of counts
that is 95% certain to contain the true average value. So long as the number of counts, C, is greater than about 50 you can
calculate the confidence interval using this approximate equation:
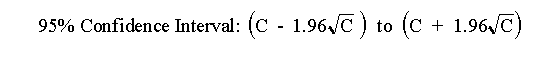
GraphPad StatMate (a program we shall use during the course) does
this calculation for you using a more exact equation that can be used for any value of C. For example, if you measure 100
radioactive counts in an interval, you can be 95% sure that the true average number of counts ranges approximately between
80 and 120 (using the equation here) or more exactly between 81.37 and 121.61 (using StatMate).
When calculating the confidence interval, you must set C equal to
the total number of counts you measured experimentally, not the number of counts per minute. Example:
You placed a radioactive sample into a scintillation counter and counted for 10 min. The counter tells you that there were
225 cpm. What is the 95% confidence interval? Since you counted for 10 min, the instrument must have detected 2250 cpm. The
95% confidence interval of this number extends from 2157 to 2343. This is the confidence interval for the number of counts
in 10 min, so the 95% confidence interval for the average number of cpm extends from 216 to 234. If you had attempted to calculate
the confidence interval using the number 225 cpm rather than 2250 (actual counts detected), you would have calculated a wider
(incorrect) interval.
The Poisson distribution explains why it is helpful to counts your
samples longer when the number of counts is small. For example, this table shows the confidence interval for 100 cpm counted
for various times. When you count for longer times, the confidence interval will be narrower.
| Counting Time |
1 minute |
10 minutes |
100 minutes |
| Counts per minute (cpm) |
100 |
100 |
100 |
| Total counts |
100 |
1000 |
10000 |
| 95% CI (in counts) |
81.4 to 121.6 |
938 to 1062 |
9804 to 10196 |
| 95% CI (in cpm) |
81.4 to 121.6 |
93.8 to 106.2 |
98.0 to 102.0 |
This graph shows percent error as a function of number of counts
(C). Percent error is defined from the width of the confidence interval divided by the number of counts:
