Competitive Binding Data with One Class of Receptors
Present and future uses of competition binding assays
It is too expensive and too technically challenging to characterize
receptors only by direct radioligand binding assays. Moreover, when studying a new drug, it is usually not feasible to radiolabel
the drug prior to understanding its receptor properties. For these reasons, competition assays are used most widely in receptor
studies. One of the interesting new approaches in drug design and development is an elegant brute force approach called combinatorial
chemistry. In this approach, one starts with several structural backbones that are felt likely to be components of a receptor
pharmacophore (the structural features of a drug that cause binding and/or activation or blockade) and does chemical reactions
that allow several different "pieces" to react in a way that hundreds of products may be formed. Rather than separating this
gemisch and testing each component, the mixture is tested, and if the mixture is found to be "positive", the active species
are purified and studied more conventionally. Such an approach dramatically increases the throughput of tested compounds.
The most commonly used test system in combinatorial chemistry is the radioreceptor assay!
What is a competitive binding curve?
Competitive binding experiments measure the binding of a single
concentration of labeled ligand in the presence of various concentrations of unlabeled ligand. Typically, the concentration
of unlabeled ligand varies over at least six orders of magnitude.
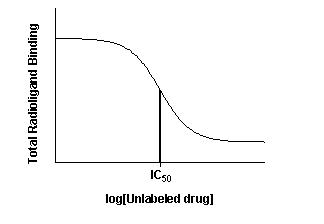
The top of the curve is a plateau at a value equal to radioligand
binding in the absence of the competing unlabeled drug. The bottom of the curve is a plateau equal to nonspecific binding.
The concentration of unlabeled drug that produces radioligand binding half way between the upper and lower plateaus is called
the IC50 (inhibitory concentration 50%) or EC50 (effective concentration 50%).
If the radioligand and competitor both bind reversibly to a single
binding site, binding at equilibrium follows this equation (where Top and Bottom are the Y values at the top and bottom plateau
of the curve).
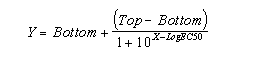
Competitive binding experiments are often used in more complicated
situations where this equation doesn't apply.
Entering data
If you performed an experiment with triplicate samples and you are
using software for analysis, let the software automatically plot the error bars. Some investigators transform their data to
percent specific binding. The problem with this approach is that you need to define how many cpm equal 100% binding and how
many equal 0% specific binding. Deciding on these values is usually somewhat arbitrary. It is usually better to enter the
data as cpm.
Decisions to make before fitting the data
Weighting
When analyzing the data, you need to decide whether to minimize
the sum of the square of the absolute distances of the points from the curve or to minimize the sum of the square of the relative
distances. The choice depends on the source of the experimental error. Follow these guidelines:
- If the bulk of the error comes from pipetting, the standard deviation
of replicate measurements will be, on average, a constant fraction of the amount of binding. In a typical experiment, for
example, the highest amount of binding might be 2000 cpm with a SD of 100 cpm. The low-est binding might be 400 cpm with a
SD of 20 cpm. With data like this, you should evaluate goodness-of-fit with relative distances. The details on how to do this
are in the next section.
- You should only consider weighting by relative distances when you
are analyzing total binding data. When analyzing specific binding (or data normalized to percent inhibi-tion), you should
evaluate goodness-of-fit using absolute distances, as there is no clear relationship between the amount of scatter and the
amount of specific binding.
Constants
With experience, you will notice that even with the use of the best
software, data that is non-ideal (i.e., most data) requires the experimenter to make decisions during the course of the analysis.
Many times, your data will span a wide range of concentrations of unlabeled drug, and clearly define the bottom or top plateaus
of the curve. If this is the case, software can estimate with accuracy the 0% and 100% values from the plateaus of the curve.
In some cases, your competition data may not define a clear bottom
plateau, but you can define the plateau from other data. All drugs that bind to the same receptor should compete all specific
radioligand binding and reach the same bottom plateau value. This means that you can define the 0% value (the bottom plateau
of the curve) by measuring radioligand binding in the presence of a standard drug at a concentration known to block all specific
binding. If you do this, make sure that you use plenty of replicates to determine this value accurately. If your definition
of the lower plateau is wrong, the values for the EC50 will be wrong as well. You can also define the top plateau as binding
in the absence of any competitor.
To summarize, if you have collected enough data to clearly define
the entire curve, let the software fit all the variables and fit the top and bottom plateaus based on the overall shape of
your data. If your data do not define a clear top or bottom plateau, you should define one or both of these values to be constants
fixed to values determined from other data. Because of factors such as intra-assay drift, this often provides a challenge
to the investigator. In the class exercises, change the top and bottom values, and observe the effects on the derived variables
and the fitted curve.
Interpreting the results
Assumptions
To interpret the results, you must make these assumptions:
- Only a small fraction of your radiolabeled ligand binds to the
tissue so that the free concentration is virtually the same as the added concentration. (There are alternative methods that
do not make this assumption.)
- There is no cooperativity.
- Your experiment has reached equilibrium.
- Binding is reversible and follows the law of mass action.
- You use a valid value for the KD of the radioligand
(e.g., under similar conditions with a similar receptor).
IC50
The IC50 is influenced by three theoretical factors (in addition
to the factors controlled, or affected, by the experimenter :
- The affinity of the receptor for the competing drug (as reflected
by its theoretical KD). If the affinity is high (i.e., small KD) the IC50 will be low.
- The concentration of the radioligand. If you choose to use a higher
concentration of radioligand, it will take a larger concentration of unlabeled drug to compete for the binding.
- The affinity of the radioligand for the receptor (KD).
It takes more unlabeled drug to compete for a tightly bound radioligand (small KD) than for a loosely bound radioligand
(high KD).
KI
- The KI is an equilibrium dissociation constant.
It is the concentration of the competing ligand that would bind to half the binding sites at equilibrium in the absence
of radioligand or other competitors.
- The subscript "I" is used to indicate that the competitor inhibited
radioligand binding. Thus, it is determined indirectly by measuring competition with a known radioligand.
- The Ki can be calculated once the IC50 is determined,
and the competition curve has been analyzed. Commonly, we use one of the equations from Cheng and Prusoff (Biochem. Pharmacol.
22: 3099-3108, 1973). Note that there is not one Cheng-Prusoff relationship, but several. The one below is for pure
competition at a single site:
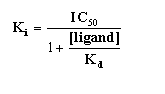
Shallow competitive binding curves
The slope factor or Hill slope
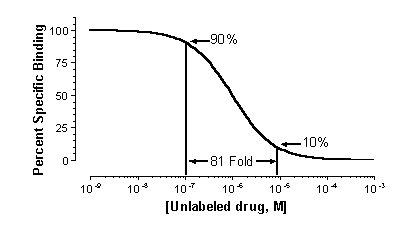
If the labeled and unlabeled ligand compete for a single binding
site, the competitive binding curve will have a shape determined by the law of mass action. In this case, the curve will descend
from 90% specific binding to 10% specific binding over an 81-fold increase in the concentration of the unlabeled drug. More
simply, virtually the entire curve will cover two log units (100-fold change in concentration).
To quantify the steepness of a competitive binding curve, one determines
a slope factor (also called Hill slope). A standard competitive binding curve that follows the law of mass action has a slope
of -1.0. If the slope is more shallow, the slope factor will be a negative fraction (i.e. -0.85 or -0.60).
The Hill Equation
The Hill Equation provides information about the nature of ligand-receptor
interactions. Widely used in the past, and often demanded by reviewers, its utility has been decreased by the wide availability
of nonlinear regression analysis programs (like Prism, a program designed for receptor analysis). Thus, while less useful,
it is important to understand the Hill equation since it has been a standard way of describing the slope factor (also called
Hill slope or Hill number). It is derived from the binding equation modified to include the situation whereby the ligand may
have multiple sites for interaction "n".
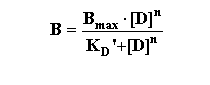
This can then be rearranged in several steps as follows:
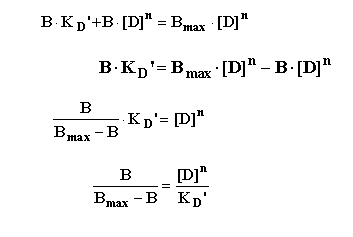
To yield the "Hill Equation".
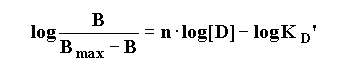
Thus, a plot of the log [Drug] vs. the logit function on the left
hand side of the equation results in a straight-line with a slope = nH. (the Hill slope or Hill number)
The slope factor describes the steepness of a curve. In most situations,
there is no way to interpret the value in terms of chemistry or biology. If the slope factor is far from 1.0, then the binding
does not follow the law of mass action with a single site. Often investigators transform the data to create a linear Hill
plot. The slope of this plot equals the slope factor, and the intercept with an ordinate value of "0" equals the IC50.
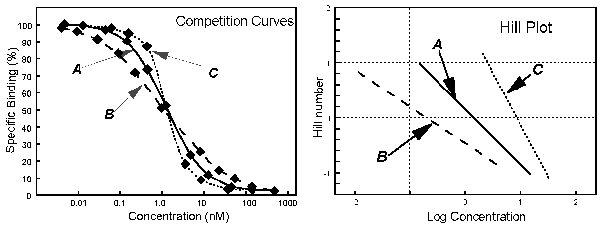
 In
these figures, A is a curve of normal steepness, with a Hill slope of 1. B is a curve of shallow steepness, with a Hill slope
1. C is a steep curve with a Hill slope 1
In
these figures, A is a curve of normal steepness, with a Hill slope of 1. B is a curve of shallow steepness, with a Hill slope
1. C is a steep curve with a Hill slope 1

Explanations for shallow binding curves include:
- Experimental problems. If the serial dilution of the unlabeled
drug concentrations was done incorrectly, the slope factor is not meaningful.
- Curve fitting problems. If the top and bottom plateaus are not
correct, then the slope factor is not meaningful. Don't try to interpret the slope factor unless the curve has clear top and
bottom plateaus.
- Negative cooperativity. You will observe a shallow binding curve
if the binding sites are clustered (perhaps several binding sites per molecule) and binding of the unlabeled ligand to one
site causes the remaining site(s) to bind the unlabeled ligand with lower affinity.
- Heterogeneous receptors. The receptors do not all bind the unlabeled
drug with the same affinity.
- Ternary complex model with limiting availability of G protein.
This is a common situation that results with heterogeneous receptors. See below.
Competitive binding with two sites
Often, data does not fit one site models well, and a two site analysis
may be useful. (Normal experimental variance usually does not permit discrimination of two from three high affinity sites).
Some assumptions needed to do such analyses include:
- There are two distinct classes of receptors. For example, a tissue
could contain a mixture of b1 and b2 adrenergic receptors.
- The unlabeled ligand has distinct affinities for the two sites.
- The labeled ligand has equal affinity for both sites.
- Binding has reached equilibrium.
- A small fraction of both labeled and unlabeled ligand bind. This
means that the concentration of labeled ligand added is very close to the free concentration in all tubes.
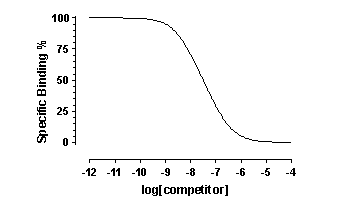
When you look at the competitive binding curve, you will only see
a biphasic curve only in the rare cases where the affinities are very different. More often you will see a shallow curve with
the two components blurred together. For example, this graph shows competition for two equally abundant sites with a ten fold
(one log unit) difference in EC50. If you look carefully, you can see that the curve is shallow (it takes more than two log
units to go from 90% to 10% competition), but you cannot see two distinct components.
Comparing one- and two-site models
Some software (like Prism) can simultaneously fit your data to two
equations and compare the two fits. This feature is commonly used to compare a one-site competitive binding model and a two-site
competitive binding model. Since the model has an extra parameter and thus the curve has an extra inflection point, the two-site
model almost always fits the data better than the one site model. And a three site model would fit even better. Before accepting
the more complicated models, you need to ask whether the improvement in goodness of fit is more than you would expect by chance.
Prism, for example, answers this question with an F test that yields a P value that answers this question: If the one site
model were really correct, what is the chance that randomly chosen data points would fit to a two-site model this much better
(or more so) than to a one-site model?
Before looking at Prism’s comparison of the two equations,
you should look at both fits yourself. Sometimes the two-site fit gives results that are clearly nonsense. Here are some examples
to when to disregard a two-site fit:
- The two IC50 values are almost identical.
- One of the IC50 values is outside the range of your data.
- The fraction of the total binding represented by any of the sites
is either close to 1.0, or to 0.0. In this case, most of the receptors have the same affinity, and the IC50 value for the
low occurring site cannot be reliable.
- The calculated values for either the BOTTOM or TOP of your curves
are far from the range of real values in your experiment.
Basic Principle: If the results don’t make sense, don’t
believe them (don’t ever say "The computer said ...."). Only pay attention to the results of the comparison when both
fits are reasonable.
This equation has five variables: the top and bottom plateau binding,
the fraction of the receptors of the first class, and the IC50 of competition of the unlabeled ligand for both classes of
receptors. If you know the KD of the labeled ligand and its concentration, you can convert the IC50 values to Ki
values.
Agonist binding and the ternary complex
If you use an antagonist radioligand and compete with an unlabeled
agonist you are likely to observe a shallow competitive binding curve. Receptors can interact with intracellular molecules
in a way that alters agonist binding. One situation that has been well studied is the competition of unlabeled agonist ligand
for antagonist radioligands for binding to receptors linked to G proteins in membrane preparations lacking GTP.
The ternary complex model explains some of these data. This model
shows binding of an agonist (A) to receptor (R) coupled to G protein (G):
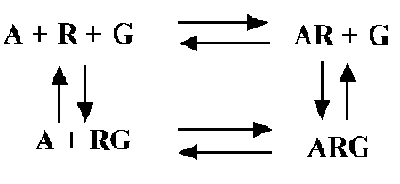
Some features of the model:
- The left half of the picture shows the equilibrium between receptor
(R) and G protein (G) in the absence of agonist (A).
- The right half shows the agonist-receptor complex binding to G
to form the ternary complex (ARG).
- The top part shows the binding of agonist to receptors not linked
to G.
- The bottom part shows the binding of agonist to receptors linked
to G.
- Not shown is the radioligand L that binds with equal affinity to
R and RG.
- This model can explain the shape of agonist competition curves
if the following are true:
- Agonist binds more tightly to the receptor-G complex than to receptor
alone. In other words, A binds with low affinity to R but to high affinity to RG.
- The total concentration of G is limiting. If G is present in higher
concentrations than R, this model does not predict shallow binding curves (Neubig et al. Mol. Pharmacol. 28:475, 1985).
- GTP is omitted from the incubation. When GTP is present, as it
is inside cells, the model is more complicated. Soon after the ARG complex forms, GTP binds to the G protein, and activates
it. The activated G then dissociates from AR, and its * subunit dissociates from the ** subunits. Because the ARG complex
is so transient, all you see in binding experiments is low affinity AR binding.
A few investigators have fit data to equations describing the ternary
complex. Most do not, for these reasons:
- The model is too complicated to fit well. There are too many parameters
to fit. You need to fit all four equilibrium constants, plus the relative concentration of receptor to G.
- The model is too simple to be useful. The ternary complex model
only predicts a shallow competitive binding curve when the total concentration of G is less than or equal to the total concentration
of receptors. But biochemical evidence in many systems demonstrates that G is present in vastly higher concentrations than
receptors. Yet these systems demonstrate shallow competitive binding curves. To resolve this discrepancy, you must make the
model still more complicated (either many of the G proteins are not available to bind to receptors, or many of the receptors
are not available to bind to G proteins). For a review of these issues, see RR Neubig, Faseb J. 8:939-946, 1994.
Rather than fit data to the ternary complex model, most investigators
fit data to a two-site binding model. The data fit well to the two site model, even though we know it is too simple. The two-site
model is shown below - it is not the same as the ternary complex model.
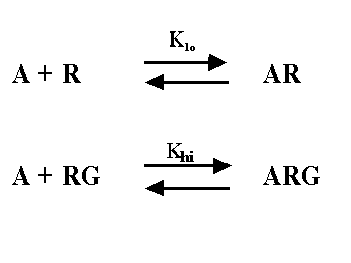
The two-site model, as its name implies, assumes that there are
two distinct kinds of receptors. In this context, one kind of receptor is coupled to G, and the other is not. This simpler
model provides values for Klo, Khi, and relative numbers of the two kinds of receptors, and these can
be compared after different treatments. Since we know that the two-site model is too simple (the ternary complex is a better
model for many systems) you shouldn't interpret Khi and Klo strictly. Nonetheless, the ratio of the
Khi and Klo is a useful empirical measure which often correlates with agonist efficacy.
This short section is hardly adequate to explain the complexities
of the ternary complex model, but should be sufficient to keep you from making common mistakes in interpretation.
Ligand, Prism and other curve fitting programs
The first widely used program in the field was "Ligand", a freely
available, special-purpose nonlinear regression program for analyzing equilibrium radioligand binding data written by Munson,
Rodbard, and collaborators (Methods in Enzymology 92:543, 1983). Today, there are many other alternatives to Ligand (for many
different computer platforms) that may be easier to use or more facile with certain types of analyses.
